Structure of the SARS-CoV spike glycoprotein in complex with a biparatopic Bicycle molecule
Pellegrino, S., Drulyte, I., Harman, M., Bezerra, G.A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike glycoprotein | 1,133 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 6 Gene Names: S, 2 | 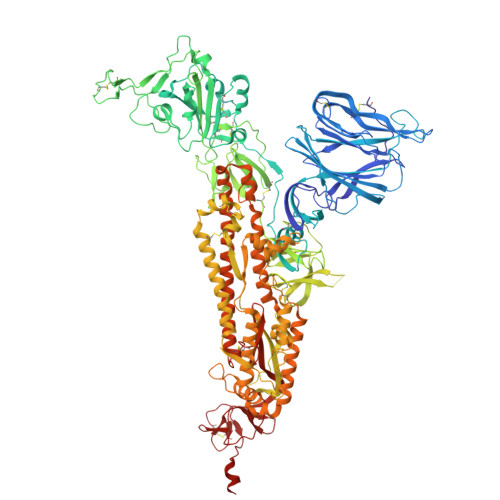 | |
UniProt | |||||
Find proteins for P0DTC2 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTC2 Go to UniProtKB: P0DTC2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTC2 | ||||
Glycosylation | |||||
| Glycosylation Sites: 15 | Go to GlyGen: P0DTC2-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Biparatopic bicycle molecule (14mer) | 15 | synthetic construct | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Biparatopic bicycle molecule (15mer) | 16 | synthetic construct | Mutation(s): 0 | 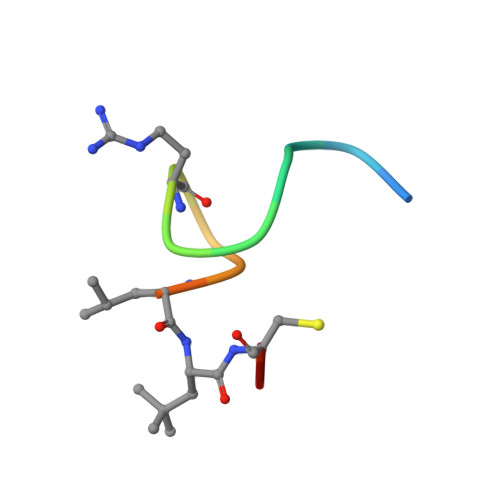 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | AA [auth CA], BA [auth CB], CA [auth CC], DA [auth CD], EA [auth CE], AA [auth CA], BA [auth CB], CA [auth CC], DA [auth CD], EA [auth CE], FA [auth CF], GA [auth CG], J [auth AA], K [auth AB], L [auth AC], M [auth AD], N [auth AE], O [auth AF], P [auth AG], Q [auth AH], R [auth BA], S [auth BB], T [auth BC], U [auth BD], V [auth BE], W [auth BF], X [auth BG], Y [auth BH], Z [auth BI] | 2 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| R06 (Subject of Investigation/LOI) Query on R06 | JB [auth D], KB [auth E], LB [auth F] | 1-[3,5-bis(2-chloranylethanoyl)-1,3,5-triazinan-1-yl]-2-chloranyl-ethanone C9 H12 Cl3 N3 O3 FIABSBGSWCLPPP-UHFFFAOYSA-N |  | ||
| STE Query on STE | IB [auth C], PA [auth A], XA [auth B] | STEARIC ACID C18 H36 O2 QIQXTHQIDYTFRH-UHFFFAOYSA-N |  | ||
| KZ0 (Subject of Investigation/LOI) Query on KZ0 | MB [auth G], NB [auth H], OB [auth I] | 2,4,6-tris(chloromethyl)-1,3,5-triazine C6 H6 Cl3 N3 RICRAVHJCLFPFF-UHFFFAOYSA-N |  | ||
| NAG Query on NAG | AB [auth C] BB [auth C] CB [auth C] DB [auth C] EB [auth C] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| 4J5 Query on 4J5 | G, H, I | L-PEPTIDE LINKING | C5 H13 N4 O2 |  | ARG |
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20.1_4487: |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other private | United Kingdom | -- |