Catalytic mechanism and differential alarmone regulation of a conserved stringent nucleosidase
Baerentsen, R.L., Kronborg, K., Brodersen, D.E., Zhang, Y.E.(2025) Structure
Experimental Data Snapshot
Starting Model: experimental
View more details
(2025) Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Pyrimidine/purine nucleotide 5'-monophosphate nucleosidase | 458 | Escherichia coli K-12 | Mutation(s): 1 Gene Names: ppnN, ygdH, b2795, JW2766 EC: 3.2.2 (PDB Primary Data), 3.2.2.10 (PDB Primary Data), 3.2.2.4 (PDB Primary Data) | 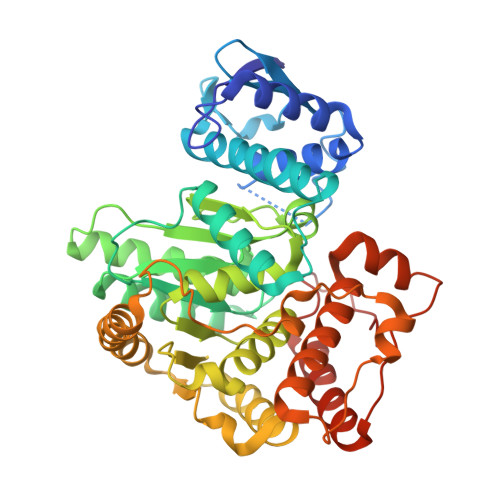 | |
UniProt | |||||
Find proteins for P0ADR8 (Escherichia coli (strain K12)) Explore P0ADR8 Go to UniProtKB: P0ADR8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ADR8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 0O2 (Subject of Investigation/LOI) Query on 0O2 | F [auth A], H [auth B], K [auth D], L [auth D] | guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) C10 H18 N5 O20 P5 KCPMACXZAITQAX-UUOKFMHZSA-N |  | ||
| HSX (Subject of Investigation/LOI) Query on HSX | E [auth A], G [auth B], I [auth C], J [auth D] | 5-O-phosphono-alpha-D-ribofuranose C5 H11 O8 P KTVPXOYAKDPRHY-AIHAYLRMSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 156.919 | α = 90 |
| b = 156.919 | β = 90 |
| c = 224.654 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Novo Nordisk Foundation | Denmark | NNF17OC0028072 |
| Novo Nordisk Foundation | Denmark | NNF18OC0030646 |