Structure of AauA, a sugar-binding protein with its substrate
Josts, I., Cottam, C., Connolly, J.P.R.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sugar-binding protein | 328 | Escherichia coli | Mutation(s): 0 Gene Names: ECs_0374 | 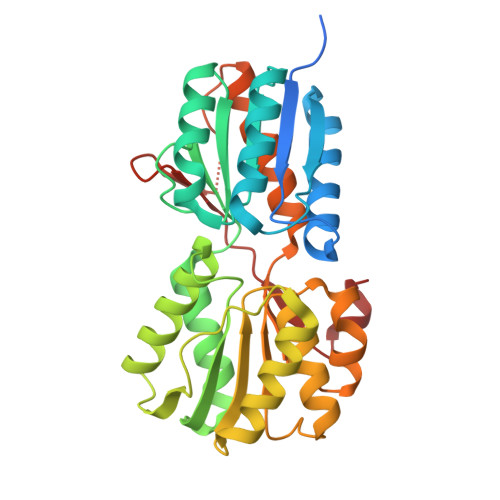 | |
UniProt | |||||
Find proteins for Q8X6A6 (Escherichia coli O157:H7) Explore Q8X6A6 Go to UniProtKB: Q8X6A6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8X6A6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PG4 Query on PG4 | B [auth A], D [auth A] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| A1IZL Query on A1IZL | C [auth A] | (2~{S},3~{R},4~{R})-2-(hydroxymethyl)oxolane-2,3,4-triol C5 H10 O5 LQXVFWRQNMEDEE-WDCZJNDASA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 76.38 | α = 90 |
| b = 66.94 | β = 94.45 |
| c = 57.85 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| xia2 | data reduction |
| Aimless | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| UK Research and Innovation (UKRI) | United Kingdom | -- |