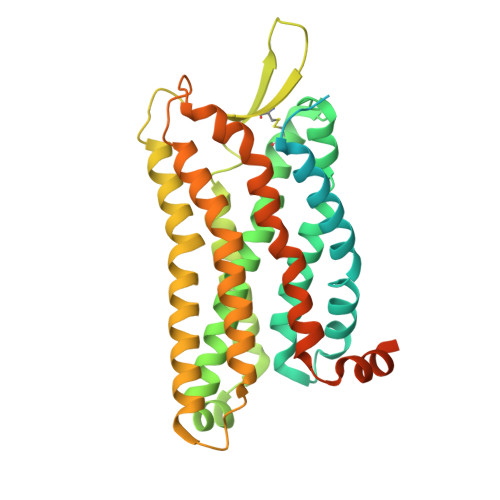
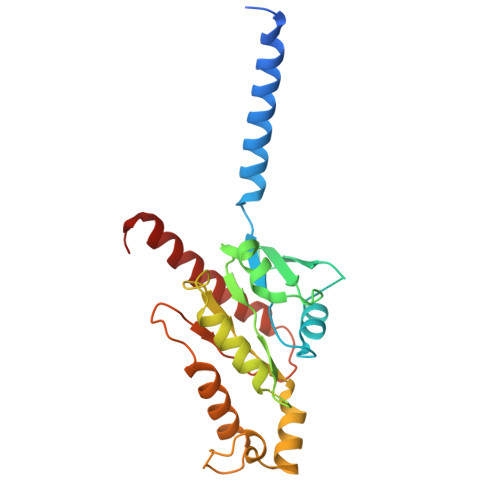
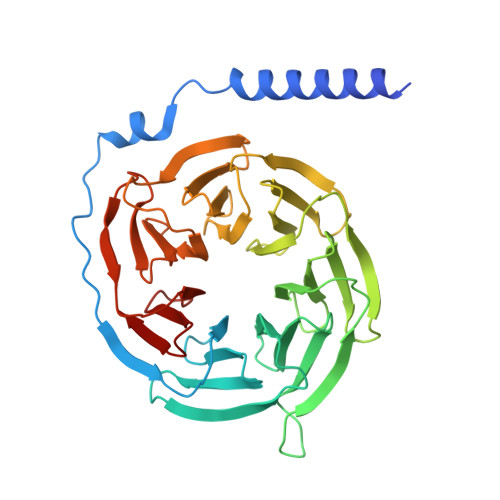
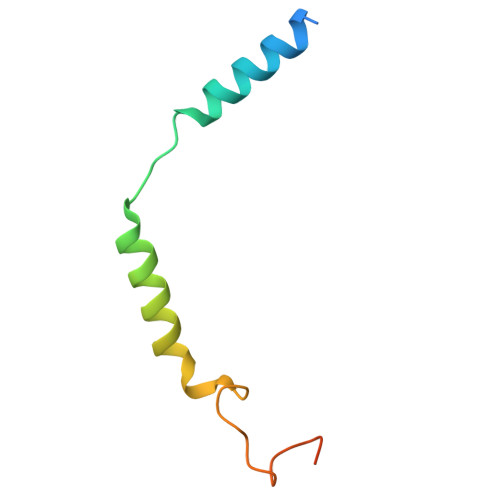
Molecular fingerprints of a convergent mechanism orchestrating diverse ligand recognition and species-specific pharmacology at the complement anaphylatoxin receptors.
Mishra, S., Yadav, M.K., Dalal, A., Ganguly, M., Yadav, R., Sawada, K., Tiwari, D., Roy, N., Banerjee, N., Fung, J.N., Marallag, J., Cui, C.S., Li, X.X., Lee, J.D., Dsouza, C.A., Saha, S., Sarma, P., Rawat, G., Zhu, H., Khant, H.A., Clark, R.J., Sano, F.K., Banerjee, R., Woodruff, T.M., Nureki, O., Gati, C., Shukla, A.K.(2025) bioRxiv
- PubMed: 40501890
- DOI: https://doi.org/10.1101/2025.05.26.656101
- Primary Citation of Related Structures:
9KUG, 9KUT, 9KV6, 9KV8, 9KVP, 9KWG, 9KWX, 9KX6, 9KXS, 9KY2, 9KYU, 9KZ2, 9KZ8, 9KZK, 9L0H, 9UMJ, 9UMR, 9UMX - PubMed Abstract:
Complement anaphylatoxin receptors (C3aR and C5aR1) are prototypical G protein-coupled receptors (GPCRs) playing crucial physiological roles in innate immunity by combating pathogenic infections and orchestrating inflammatory responses. They continue to be important therapeutic targets for multiple disorders including autoimmune diseases, acute and chronic inflammation, and allergy-related conditions. Recent structural coverage has provided important insights into their activation and signaling, however, confounding observations in the literature related to ligand efficacy and functional responses, especially in different model systems, present a major challenge for drug discovery efforts. Here, we systematically and comprehensively profile a broad set of natural and synthetic ligands at C3aR and C5aR1 and discover a previously unanticipated level of functional specialization in terms of species-specific pharmacology and receptor activation. Taking a lead from this, we determine seventeen cryo-EM structures of different ligand-receptor-G-protein complexes and uncover distinct orientation of agonists between the human and mouse receptors despite an overlapping positioning in the orthosteric binding pocket. Combined with extensive mutagenesis and functional assays, these structural snapshots allow us to decode and validate a convergent molecular mechanism involving a "Five-Point-Switch" in these receptors that orchestrates the recognition and efficacy of diverse agonists. We also identify species-specific differences at the level of phosphorylation patterns encoded in the carboxyl-terminus of these receptors and directly visualize their impact on βarr binding and activation using cryo-EM structures. Interestingly, we observe that βarrs engage with the mouse C5aR1 using a variation of previously discovered P-X-P-P phosphorylation motif via a "Sliding-Mechanism" and also exhibit distinct oligomeric state for the human vs. mouse receptors. Taken together, this study elucidates functional specialization at the complement anaphylatoxin receptors and underlying molecular mechanisms, offering a previously lacking framework with direct and immediate implications for the development of novel therapeutics.
- Department of Biological Sciences and Bioengineering, Indian Institute of Technology, Kanpur 208016, India.
Organizational Affiliation: