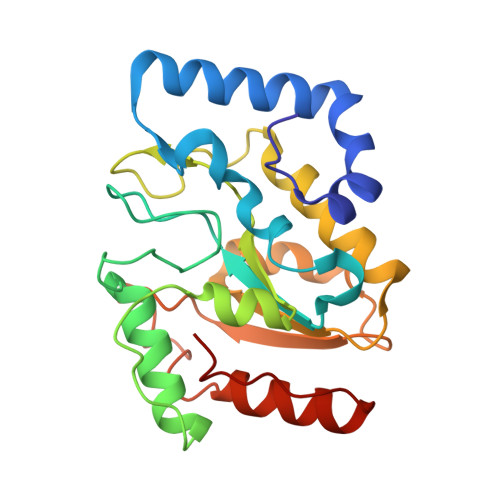
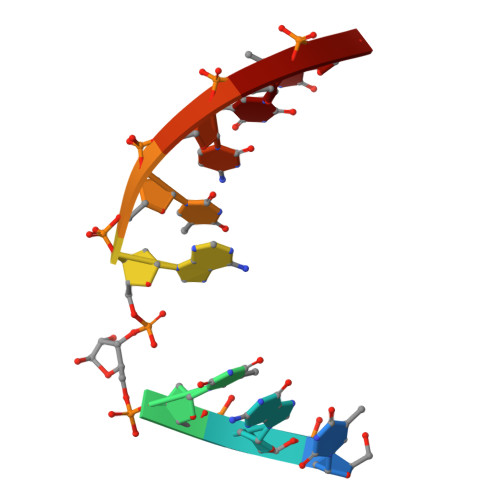
Uridine Embedded within DNA is Repaired by Uracil DNA Glycosylase via a Mechanism Distinct from That of Ribonuclease H2.
Fan, C., Zhan, X., Guo, F., Li, Q., Lu, K., Shan, X., Zhou, Y., Ren, M., Greenberg, M.M., Liu, Y., Zhou, C.(2025) J Am Chem Soc 147: 11574-11583
- PubMed: 40130361
- DOI: https://doi.org/10.1021/jacs.5c01436
- Primary Citation of Related Structures:
9LNP, 9LNQ - PubMed Abstract:
Uridine (rU) and 2'-deoxyuridine (dU) are common DNA lesions. dU is repaired through a base excision repair (BER) pathway initiated by uracil DNA glycosylase (UDG), while rU is typically removed from DNA via ribonucleotide excision repair, mediated by RNase H2. In this study, we report that rU is also repaired through the UDG-mediated BER pathway. We found that UDG catalyzes the removal of uracil from rU embedded in DNA, but exhibits no activity toward rU in RNA. Biochemical and crystallographic analyses revealed that the 2'-OH group of rU is effectively accommodated by UDG and directly participates in catalyzing the hydrolysis of the N -glycosidic bond. The abasic site product generated upon removal of uracil from rU by UDG is further processed by downstream BER enzymes to restore undamaged DNA. Our findings suggest that UDG-initiated BER constitutes a previously unrecognized pathway for the repair of rU-specific ribonucleotides. Additionally, we developed a method for selectively quantifying rU content in DNA. Using this method, we determined that rU repair by UDG is not a major pathway in human cells. This discovery expands our understanding of the diverse biological functions of UDG, and inspires further investigation to determine the role of its rU-removal in cells.
- State Key Laboratory of Elemento-Organic Chemistry, Frontiers Science Center for New Organic Matter, Department of Chemical Biology, College of Chemistry, Nankai University, Tianjin 300071, China.
Organizational Affiliation: