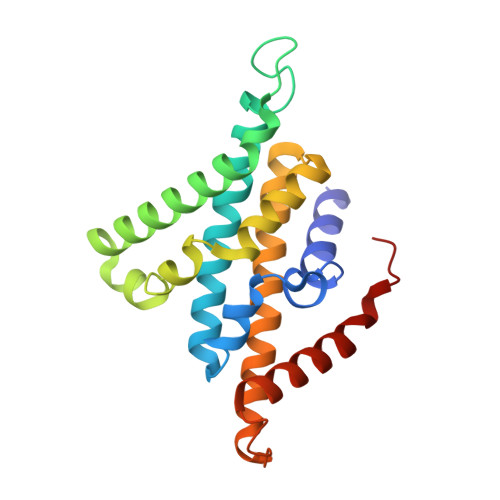
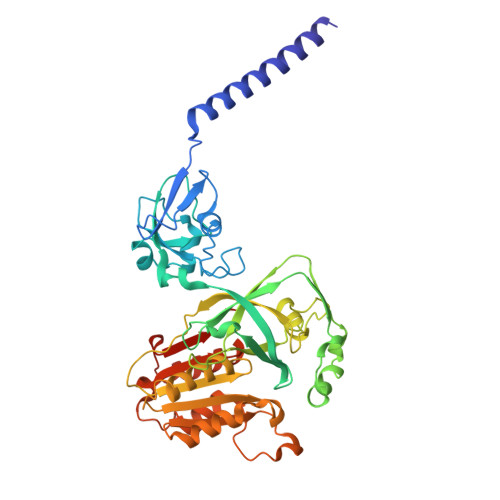
Structural Elucidation of the Mechanism for Inhibitor Resistance in the Na + -Translocating NADH-Ubiquinone Oxidoreductase from Vibrio cholerae.
Ishikawa-Fukuda, M., Kishikawa, J.I., Masuya, T., Ito, T., Butler, N.L., McFee, D., Kato, T., Barquera, B., Miyoshi, H., Murai, M.(2025) Biochemistry 64: 1963-1972
- PubMed: 40263754
- DOI: https://doi.org/10.1021/acs.biochem.5c00069
- Primary Citation of Related Structures:
9LRR - PubMed Abstract:
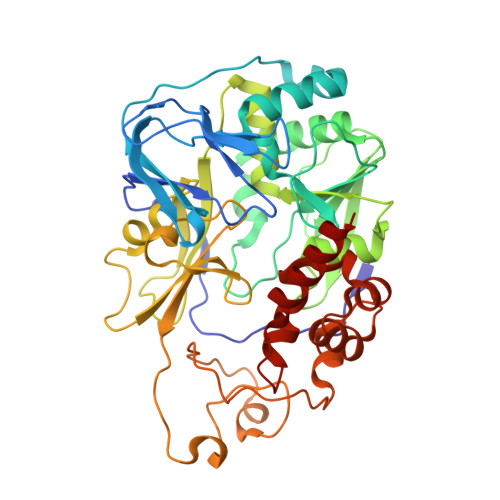
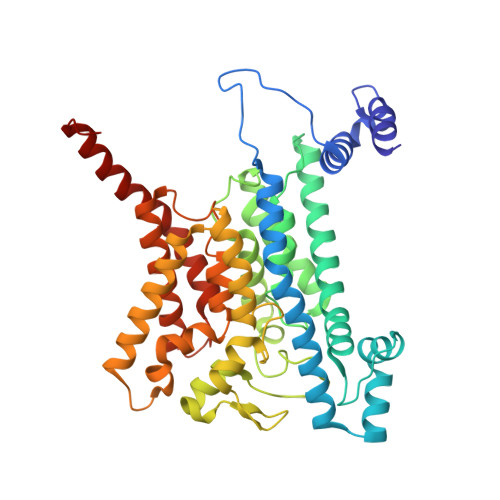
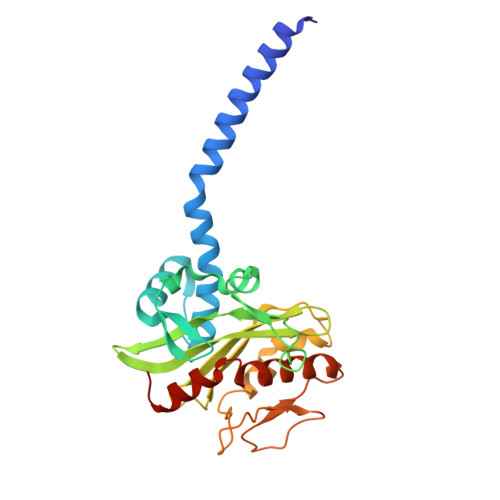
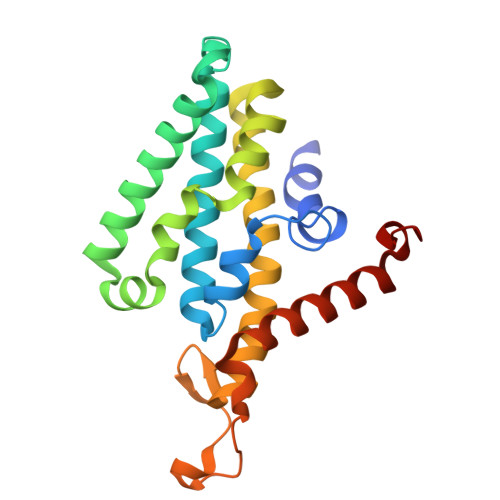
Na + -translocating NADH-ubiquinone oxidoreductase (Na + -NQR) is a unique redox-driven Na + -pump. Since this enzyme is exclusively found in prokaryotes, including the human pathogens Vibrio cholerae and Neisseria gonorrhoeae , it is a promising target for highly selective antibiotics. Korormicin A, a natural product, and a specific and potent inhibitor of V. cholerae Na + -NQR, may become a lead compound for the relevant drug design. We previously showed that the G141A mutation in the NqrB subunit (NqrB-G141A) confers moderate resistance to korormicin A (about 100-fold). However, the efficiency of photoaffinity labeling of the mutant enzyme by a photoreactive korormicin derivative was the same as in the wild-type enzyme. Because of these apparently conflicting results, the molecular mechanism underlying the korormicin A-resistance remains elusive. In the present study, we determined the cryo-EM structure of the V. cholerae NqrB-G141A mutant in the presence of bound korormicin A, and compared it to the corresponding structure from the wild-type enzyme. The toxophoric moiety of korormicin A binds to the mutant enzyme similarly to how it binds to the wild type. However, the added bulk of the alanine-141 excludes the alkyl side chain from the binding cavity, resulting in a decrease in the binding affinity. In fact, isothermal titration calorimetry revealed that the binding affinity of korormicin to the NqrB-G141A mutant is significantly weaker compared to the wild-type. Altogether, we conclude that the inhibitory potency of korormicin A is weaker in the NqrB-G141A mutant due to the decrease in its binding affinity to the altered binding cavity.
- Division of Applied Life Sciences, Graduate School of Agriculture, Kyoto University, Kyoto 606-8502, Japan.
Organizational Affiliation: