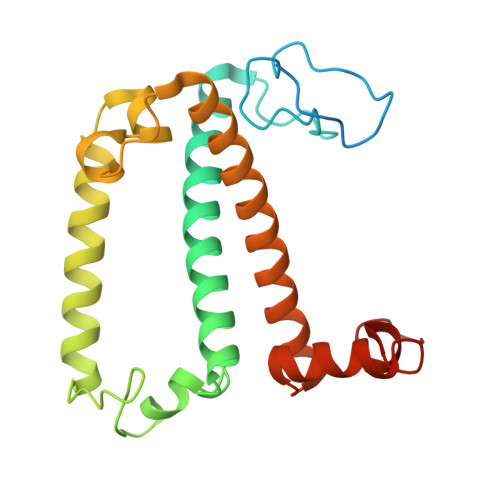
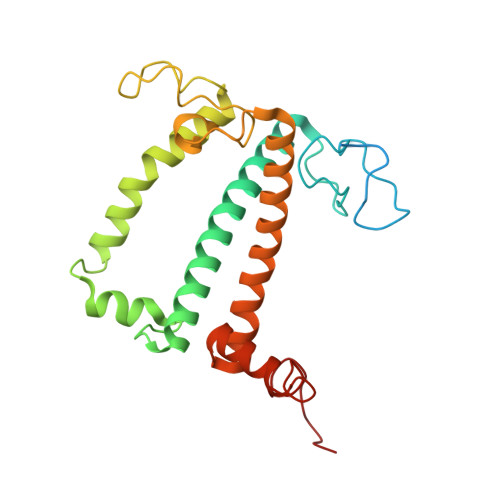
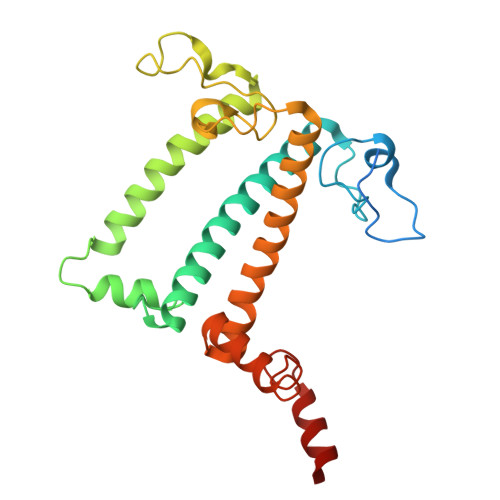
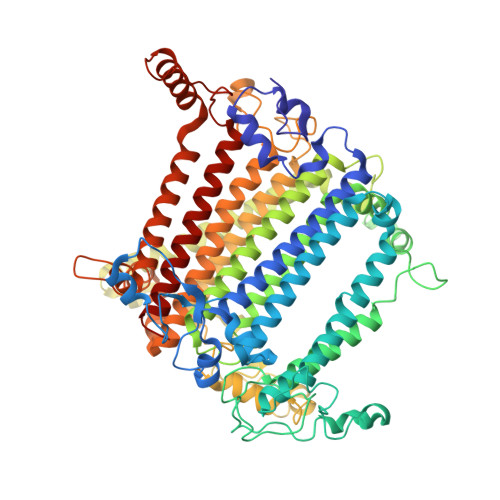
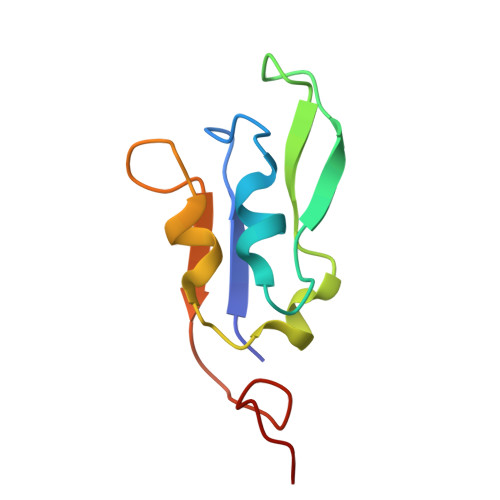
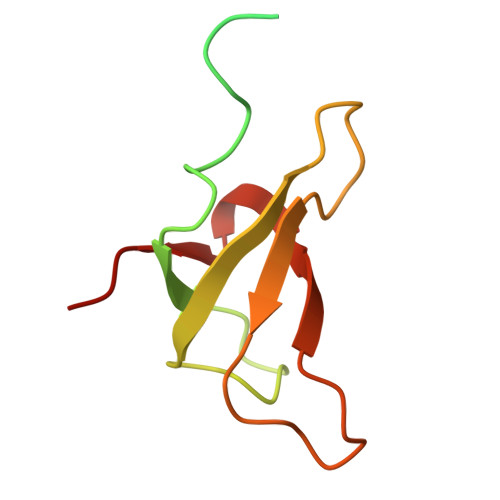
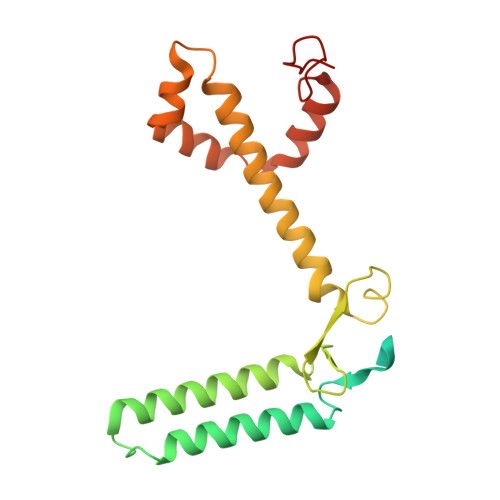
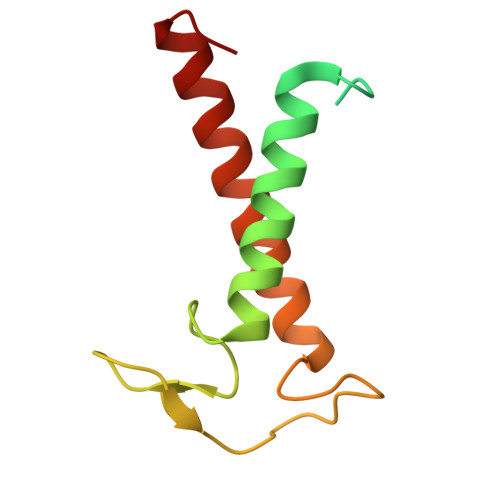
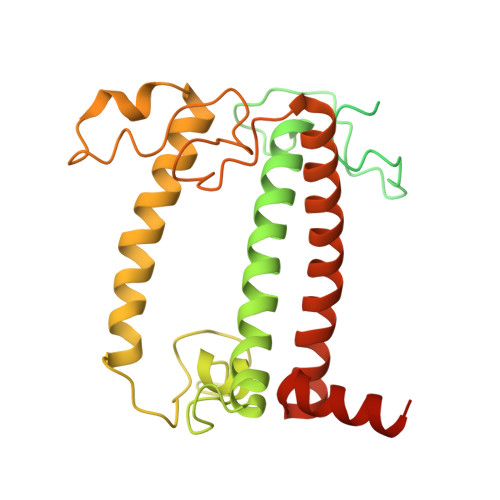
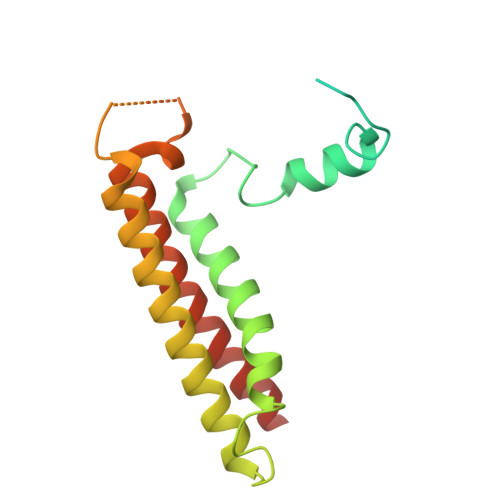
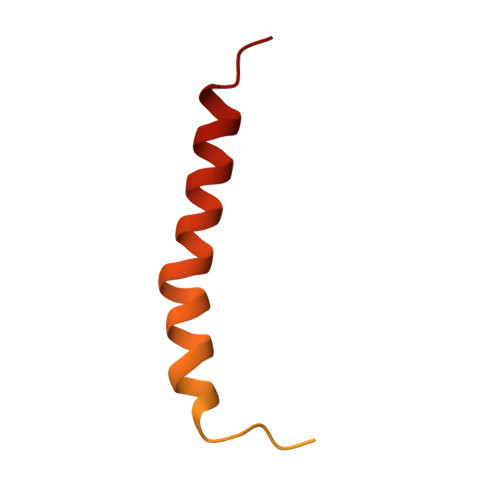
A distinct LHCI arrangement is recruited to photosystem I in Fe-starved green algae.
Liu, H.W., Khera, R., Grob, P., Gallaher, S.D., Purvine, S.O., Nicora, C.D., Lipton, M.S., Niyogi, K.K., Nogales, E., Iwai, M., Merchant, S.S.(2025) Proc Natl Acad Sci U S A 122: e2500621122-e2500621122
- PubMed: 40523173
- DOI: https://doi.org/10.1073/pnas.2500621122
- Primary Citation of Related Structures:
9MGW, 9MGZ, 9MH0, 9MH1 - PubMed Abstract:
Iron (Fe) availability limits photosynthesis at a global scale where Fe-rich photosystem (PS) I abundance is drastically reduced in Fe-poor environments. We used single-particle cryoelectron microscopy to reveal a unique Fe starvation-dependent arrangement of light-harvesting chlorophyll (LHC) proteins where Fe starvation-induced TIDI1 is found in an additional tetramer of LHC proteins associated with PSI in Dunaliella tertiolecta and Dunaliella salina . These cosmopolitan green algae are resilient to poor Fe nutrition. TIDI1 is a distinct LHC protein that co-occurs in diverse algae with flavodoxin (an Fe-independent replacement for the Fe-containing ferredoxin). The antenna expansion in eukaryotic algae we describe here is reminiscent of the iron-starvation induced ( isiA- encoding) antenna ring in cyanobacteria, which typically co-occurs with isiB , encoding flavodoxin. Our work showcases the convergent strategies that evolved after the Great Oxidation Event to maintain PSI capacity.
- Department of Plant and Microbial Biology, University of California, Berkeley, CA 94720.
Organizational Affiliation: