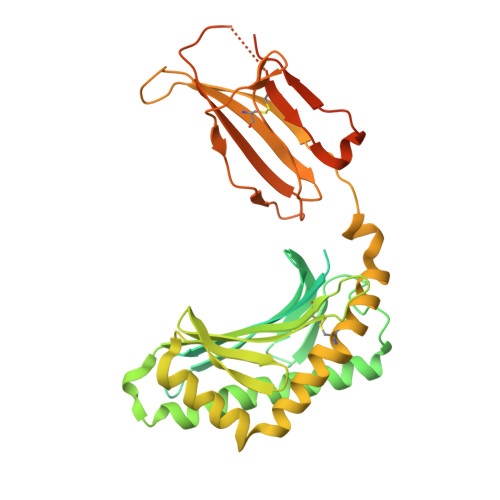
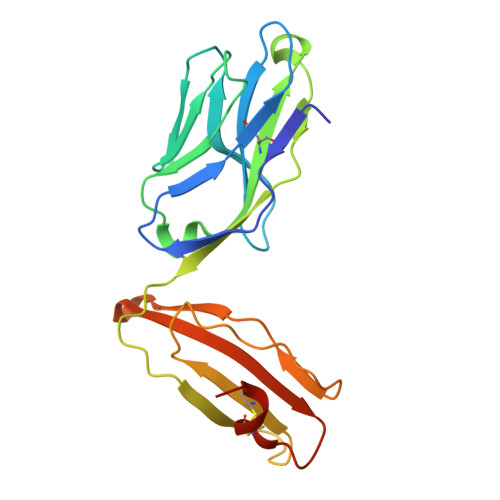
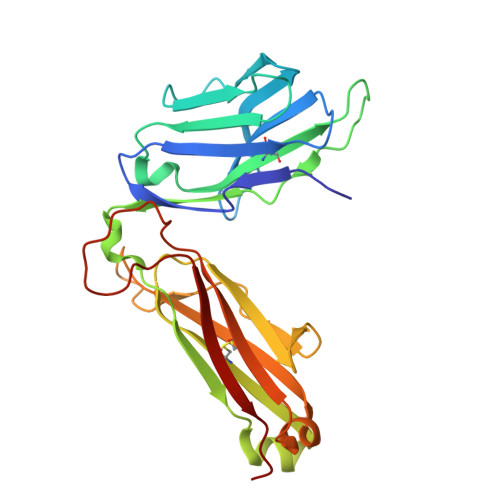
A CD1c lipid agnostic T cell receptor bispecific engager redirects T cells against CD1c + cells.
Szoke-Kovacs, R., Khakoo, S., Rangel, V.L., Della Cristina, P., Pentier, J., Khanolkar, R., El-Ajouz, S., Simmons, R., Cole, D.K., Gogolak, P., Salio, M., Karuppiah, V.(2025) Front Immunol 16: 1614610-1614610
- PubMed: 40777002
- DOI: https://doi.org/10.3389/fimmu.2025.1614610
- Primary Citation of Related Structures:
9QWJ, 9QWK - PubMed Abstract:
Immunotherapy is emerging as an efficacious treatment for some cancers, complementing traditional chemo-radiation therapies. Specific markers at the cell surface of cancer cells can be used as immunotherapy targets. However, many of these markers are defined by a patient's genetic background, limiting their use across the human population. Here, we investigated the non-polymorphic antigen presenting molecule, CD1c, that is only expressed on subsets of mature hematopoietic cells, as a potential immunotherapy target with reduced risk of off-tumor on-target toxicity in healthy tissues. We identified a T cell receptor (TCR) which recognises CD1c in a lipid independent manner and determined the crystal structure of the TCR-CD1c complex which revealed flexibility around the lipid binding region, and a new binding mechanism of auto-antigen recognition. We generated affinity enhanced variants of the TCR and fused them to an anti-CD3 antibody for T cell redirection. Lipidomic analysis revealed promiscuous lipid recognition of CD1c by the affinity enhanced TCR variants, with preference for larger lipid head group, a finding which is supported by the crystal structure. The bispecific molecule induced potent re-directed T cell killing of CD1c positive cell lines. These proof-of-concept findings demonstrate that CD1c targeting TCR bispecific engagers might be good candidates for the development of non-MHC restricted, universal therapeutics for the treatment of CD1c+ leukemias.
- Experimental Immunology, Immunocore Ltd, Abingdon, United Kingdom.
Organizational Affiliation: