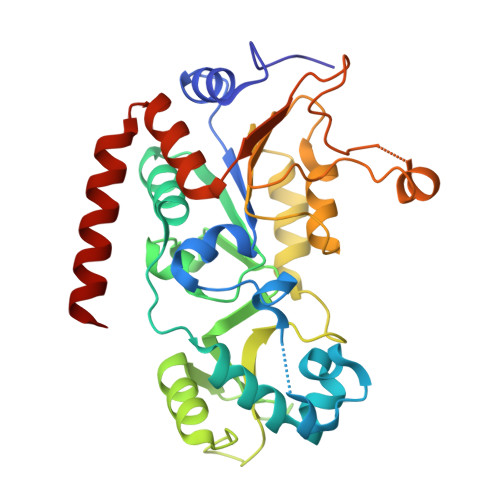
Tailored SirReal-type inhibitors enhance SIRT2 inhibition through ligand stabilization and disruption of NAD + co-factor binding.
Wirawan, R., Frei, M., Heider, A., Papenkordt, N., Friedrich, F., Wein, T., Jung, M., Groll, M., Huber, E.M., Bracher, F.(2025) RSC Med Chem
- PubMed: 40919320
- DOI: https://doi.org/10.1039/d5md00144g
- Primary Citation of Related Structures:
9S44, 9S46, 9S48 - PubMed Abstract:
Human sirtuin 2 (SIRT2) is an NAD + dependant enzyme that has been linked to the pathogenesis of various diseases, making it a promising target for pharmaceutical intervention. This study presents a systematic investigation on the inhibitory effects of SIRT2 inhibitors functionalized with diverse electrophilic functional groups. Guided by initial docking studies, we designed and synthesised 14 derivatives of two published potent lead structures 24a and SirReal2. The most potent and subtype selective SIRT2 inhibitor 29 (RW-78) exhibits an IC 50 of 26 nM, which outperforms its lead structure 24a (IC 50 = 79 nM) by a factor of 3. The increased potency of 29 is explained by halogen-π interactions with SIRT2 residues as visualized by X-ray crystallography. Furthermore, 29 interferes with NAD + binding, highlighting co-factor displacement as a valid strategy to inhibit SIRT2. Additionally, we showed cellular target engagement via NanoBRET assays in HEK293T cells (EC 50 = 15 nM). Altogether our findings provide a deeper insight into the structure-activity relationships of these SirReal-type inhibitors and offer new avenues for optimisation of SIRT2 inhibitors.
- Department of Pharmacy, Ludwig-Maximilians University Munich Butenandtstraße 5-13 81377 Munich Germany franz.bracher@cup.uni-muenchen.de.
Organizational Affiliation: