Cryo-EM structures of GnRHR: Foundations for next-generation therapeutics.
Shen, S., He, X., Liu, H., Hu, W., Xu, H.E., Duan, J.(2025) Proc Natl Acad Sci U S A 122: e2500112122-e2500112122
- PubMed: 40523184
- DOI: https://doi.org/10.1073/pnas.2500112122
- Primary Citation of Related Structures:
9U4W, 9U4Y - PubMed Abstract:
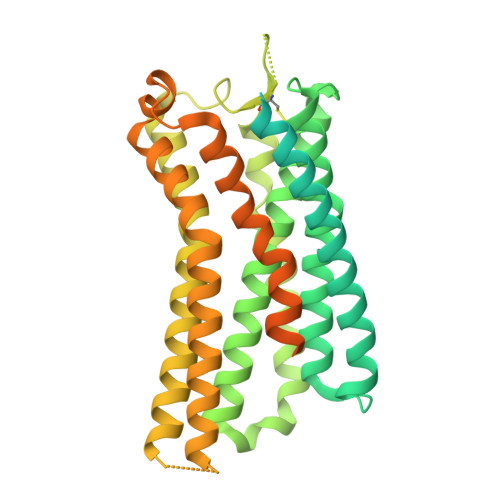
Gonadotropin-releasing hormone receptor (GnRHR) is critical for reproductive health and a key therapeutic target for endocrine disorders and hormone-responsive cancers. Using high-resolution cryoelectron microscopy, we determined the structures of Sus scrofa and Xenopus laevis GnRHRs bound to mammal GnRH, uncovering conserved and species-specific mechanisms of receptor activation and G protein coupling. The conserved "U"-shaped GnRH conformation mediates high-affinity binding through key interactions with residues such as K 3.32 , Y 6.51 , and Y 6.52 . Species-specific variations in extracellular loops and receptor-ligand contacts fine-tune receptor function, while ligand binding induces structural rearrangements, including N terminus displacement and TM6 rotation, critical for signaling. Structure-activity relationship analysis demonstrates how D-amino acid substitutions in GnRH analogs enhance stability and receptor affinity. Distinct binding modes of agonists and antagonists elucidate mechanisms of ligand-dependent activation and inactivation. These insights lay the groundwork for designing next-generation GnRHR therapeutics with enhanced specificity and efficacy for conditions like endometriosis, prostate cancer, and infertility.
- State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China.
Organizational Affiliation: