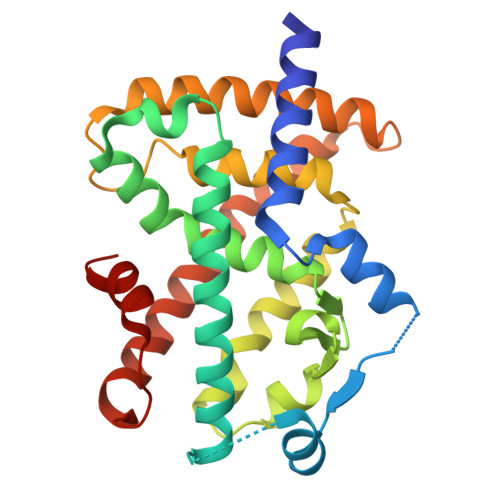
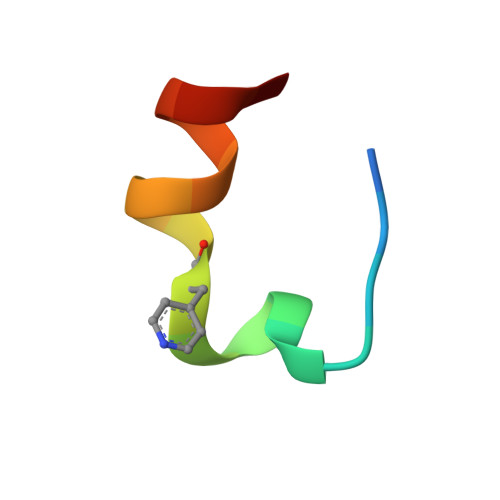De Novo Discovery of alpha , alpha-Disubstituted alpha-amino Acid-Containing alpha-helical Peptides as Competitive PPAR gamma PPI Inhibitors.
Sigal, M., Egner, M., Okada, C., Merk, D., Sengoku, T., Katoh, T., Suga, H.(2025) J Am Chem Soc
- PubMed: 41188059
- DOI: https://doi.org/10.1021/jacs.5c13803
- Primary Citation of Related Structures:
9V8D, 9V8E, 9V8F, 9V8G, 9V8H - PubMed Abstract:
α,α-Disubstituted α-amino acids (dαAAs) are important building blocks for peptidomimetics as they are strong inducers of helicity and protect against proteolytic degradation. However, de novo discovery of dαAA-containing peptides with genetically encoded libraries is limited due to their poor incorporation efficiency. Here, we report the optimized ribosomal incorporation of multiple achiral dαAAs into peptide libraries and their application to high-throughput (>10 12 members) affinity selection against the nuclear receptor peroxisome proliferator-activated receptor-gamma (PPARγ). This dαAA-based screening methodology discovered potent linear and macrocyclic α-helical peptides with low-to-sub nanomolar binding affinities. Hit peptides were proteolytically stable in serum and cell permeable, allowing for in cellulo antagonism of PPARγ. X-ray crystallography revealed that dαAA-containing peptides bound at the α-helical protein-protein interaction (PPI) interface via an α-helical conformation. This work validates the potential of a dαAA-based, α-helical discovery platform, providing access to new chemical and conformational space to de novo identify novel α-helical peptidomimetics.
- Department of Chemistry, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan.
Organizational Affiliation: