Nuclear pores dilate and constrict in cellulo
Christian E. Zimmerli, Matteo Allegretti, Vasileios Rantos, Sara K. Goetz, Agnieszka Obarska-Kosinska, Ievgeniia Zagoriy, Aliaksandr Halavatyi, Gerhard Hummer, Julia Mahamid, Jan Kosinski, Martin Beck(2021) Science
- PubMed: 34762489
- DOI: https://doi.org/10.1126/science.abd9776
- Primary Citation of Related Structures:
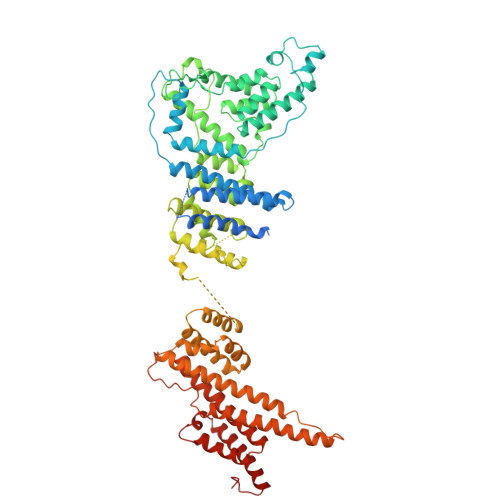
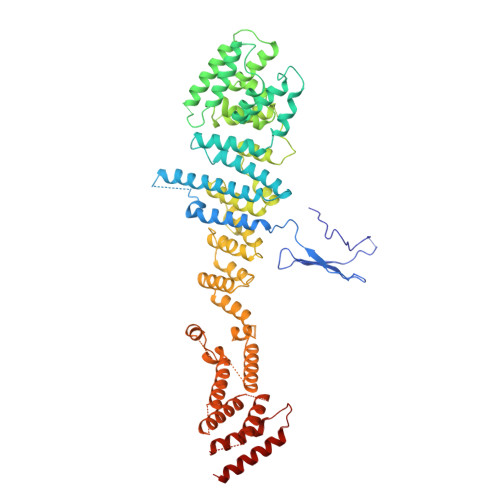
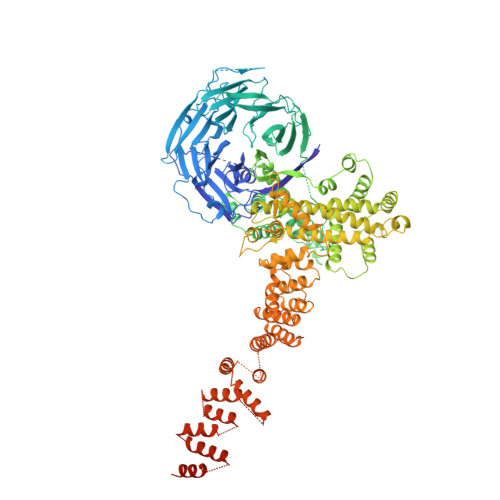
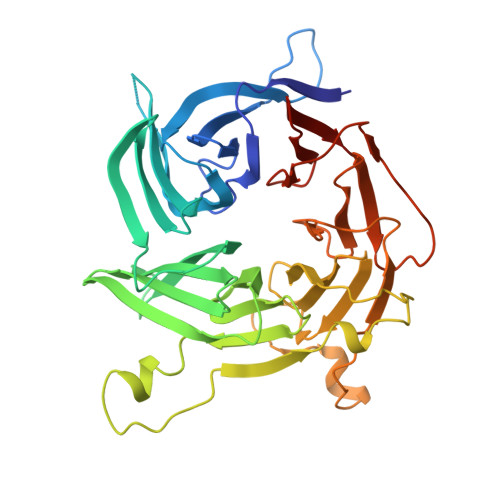
9A1M, 9A1N, 9A1O - PubMed Abstract:
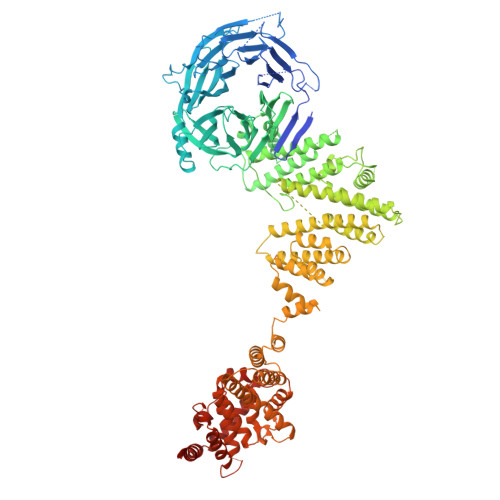
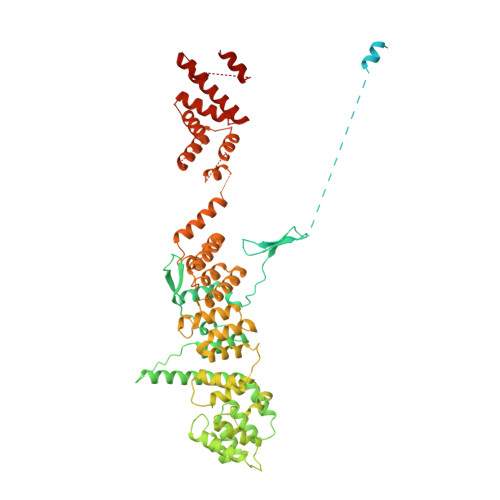
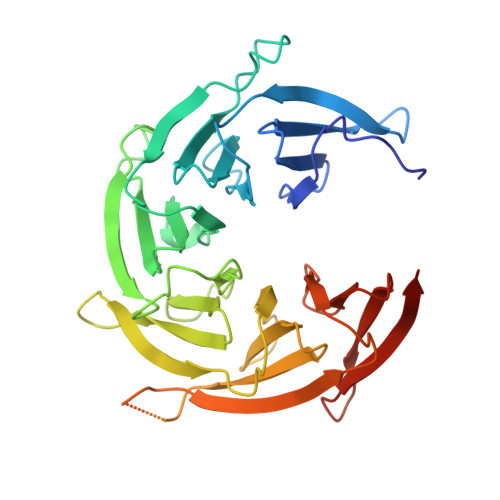
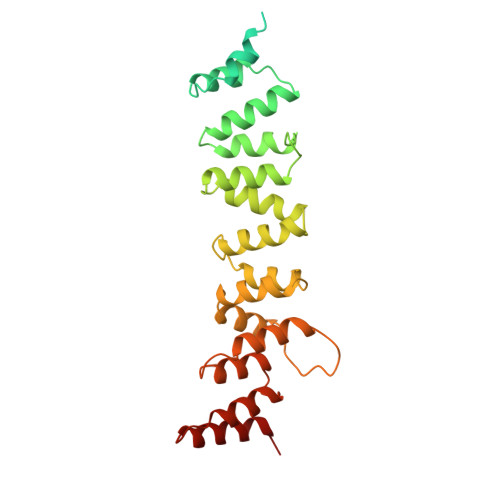
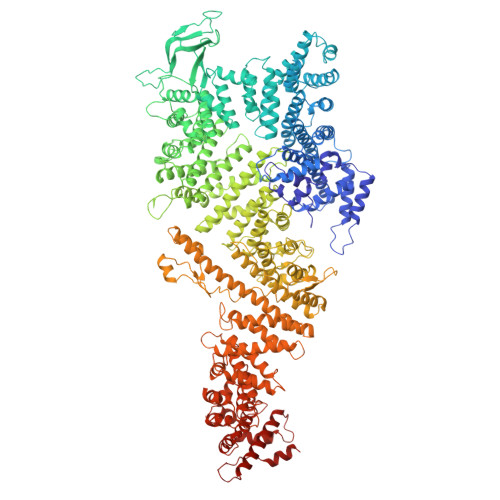
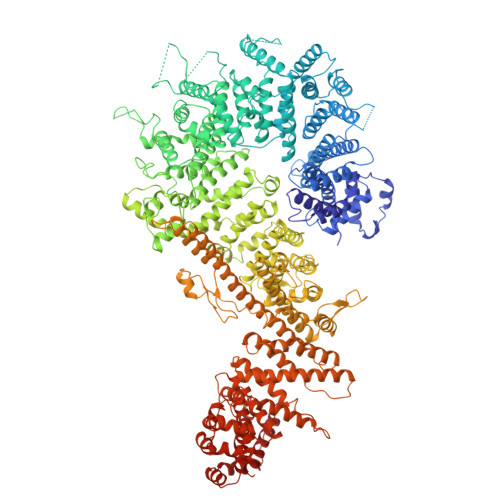
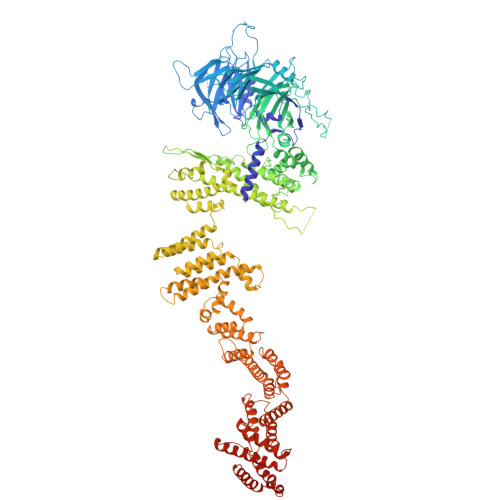
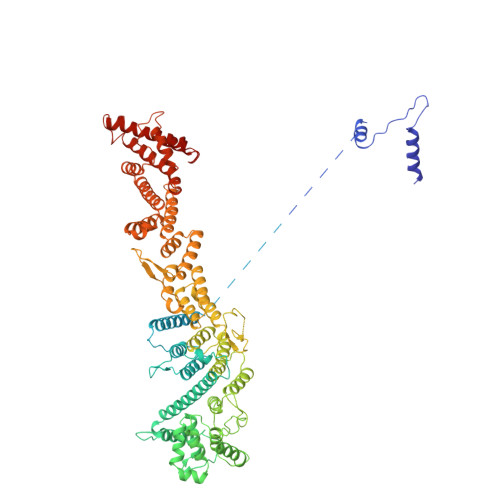
In eukaryotic cells, nuclear pore complexes (NPCs) fuse the inner and outer nuclear membranes and mediate nucleocytoplasmic exchange. They are made of 30 different nucleoporins and form a cylindrical architecture around an aqueous central channel. This architecture is highly dynamic in space and time. Variations in NPC diameter have been reported, but the physiological circumstances and the molecular details remain unknown. Here, we combined cryo–electron tomography with integrative structural modeling to capture a molecular movie of the respective large-scale conformational changes in cellulo. Although NPCs of exponentially growing cells adopted a dilated conformation, they reversibly constricted upon cellular energy depletion or conditions of hypertonic osmotic stress. Our data point to a model where the nuclear envelope membrane tension is linked to the conformation of the NPC.
- Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), 69117 Heidelberg, Germany.
Organizational Affiliation: